Results and Conclusions
Results
Regression
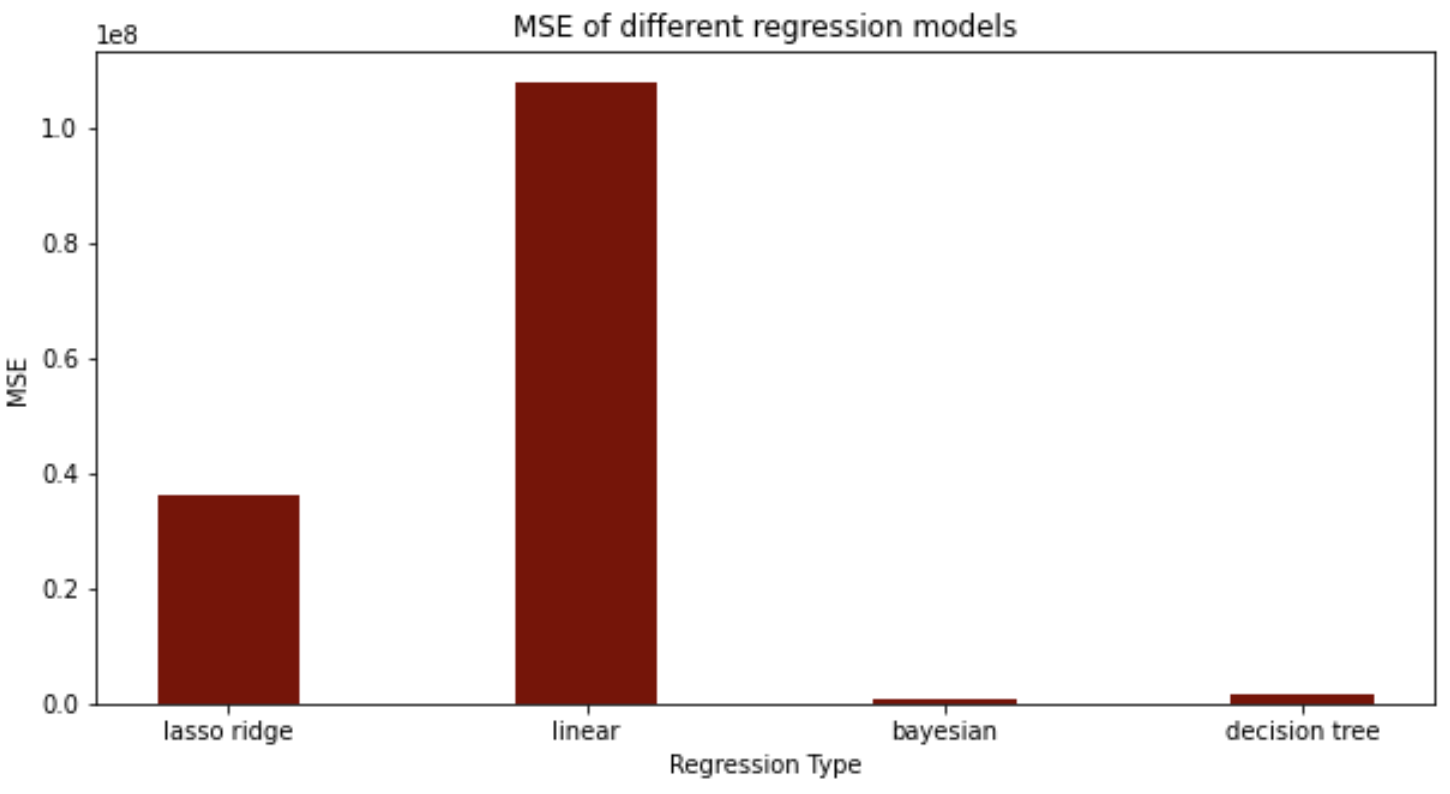
For our regression model, we found that the bayesian and decision tree models performed much better than the lasso ridge regression and linear regression, when comparing the mean squared errors.
Classification
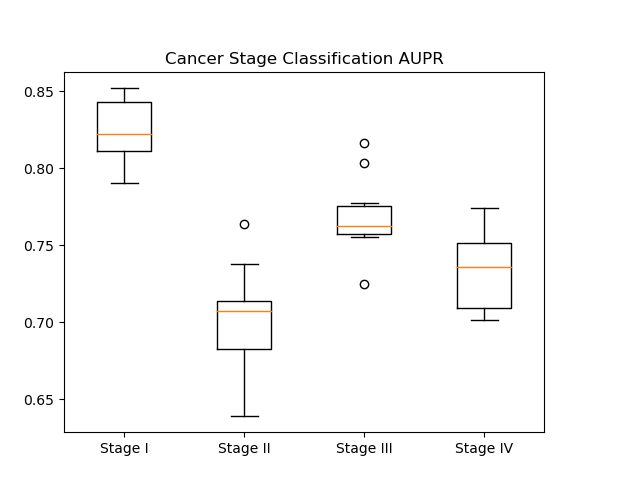
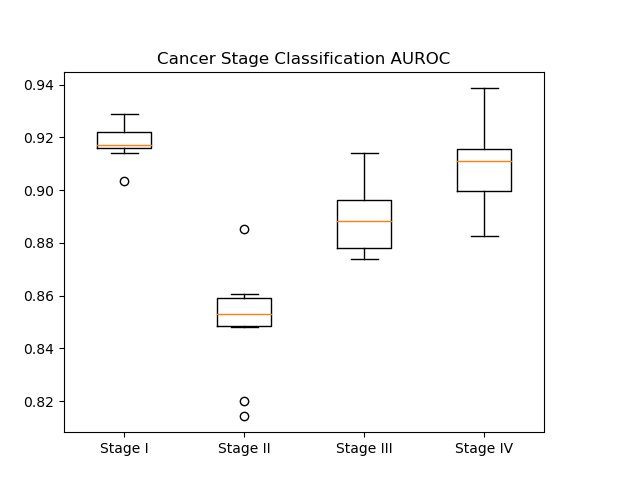
In our results, we generated AUROC and AUPR plots for our classification of cancer stage. AUROC is the area under the receiver operator characteristic curve, which essentially shows our true positive rate. The AUPR plot, or the area under the precision recall curve shows the precision of our classifier.
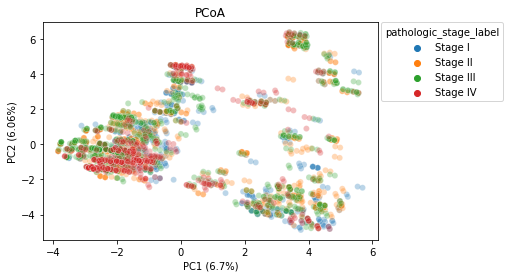
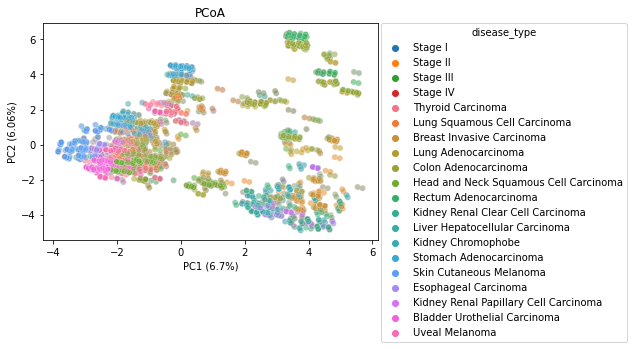
PCoA
We generated two Principal Coordinate Analysis plots, showing the separation between stages as well as the separation between disease types by a euclidean distance metric. We can see some similar clustering when comparing the two plots, indicating there likely being a relation between the cancer stage and the specific cancer type. This could be a confounding caused by the way that the data was collected, and may be something to explore in further research if there is more data.
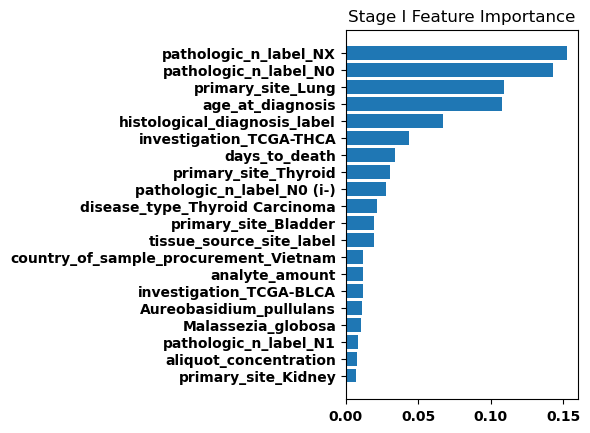
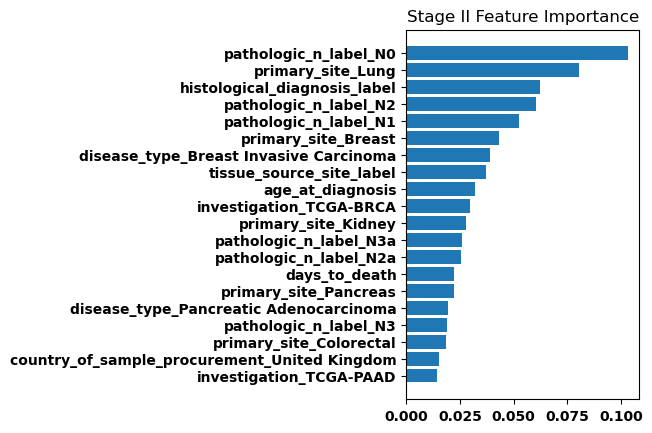
Feature Weights
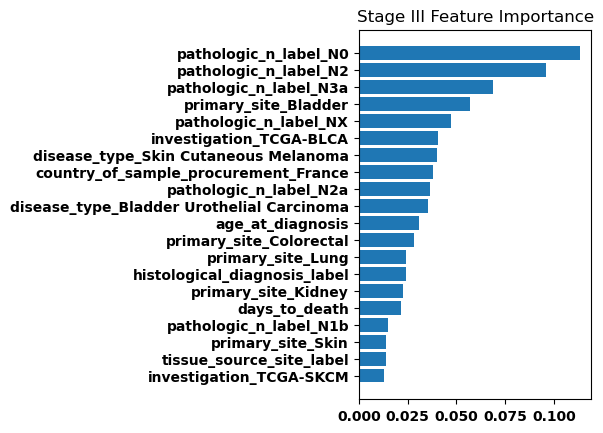
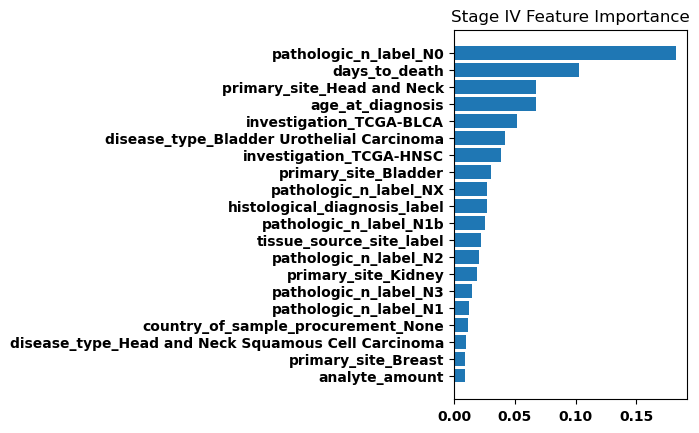
Finally, we generated bar plots showing the features that had the greatest importance in our classification model. Overall the feature importance for all the stages was relatively similar, but there were some differences especially when comparing between stage I and stage IV.
Discussion
Our study successfully achieved a high level of accuracy in classifying the stage of various cancer tumors using a combination of metadata and counts data. Notably, the inclusion of metadata in our model increased model performance compared to the original study. However, the features that our model identified as most important did not include any microbial features. It is possible that the microbial features each had a relatively small effect on the model, making them less significant than the metadata. Future studies may want to investigate methods to boost the impact of microbial features.
Reducing the number of cancer stages to four may have contributed to our model’s performance by reducing the risk of inaccuracy in attempting to classify too many stages.
Additionally, our regression model for predicting days to death was a novel concept not attempted in the original study. Despite utilizing only metadata and counts data, we achieved respectable accuracy levels in our predictions.